Research
Our research aims to understand how our genome is decoded to allow proper developmental fate transitions during embryogenesis, with a focus on epigenetic mechanisms. To address these questions, we use developing zebrafish embryos as a powerful model system. We use a combination of wet lab and dry lab approaches, including morpholino injection, immunofluorescence and imaging, epigenomic profiling, next-generation sequencing, bioinformatics and computational analyses.
Epigenetic control of gene expression programs
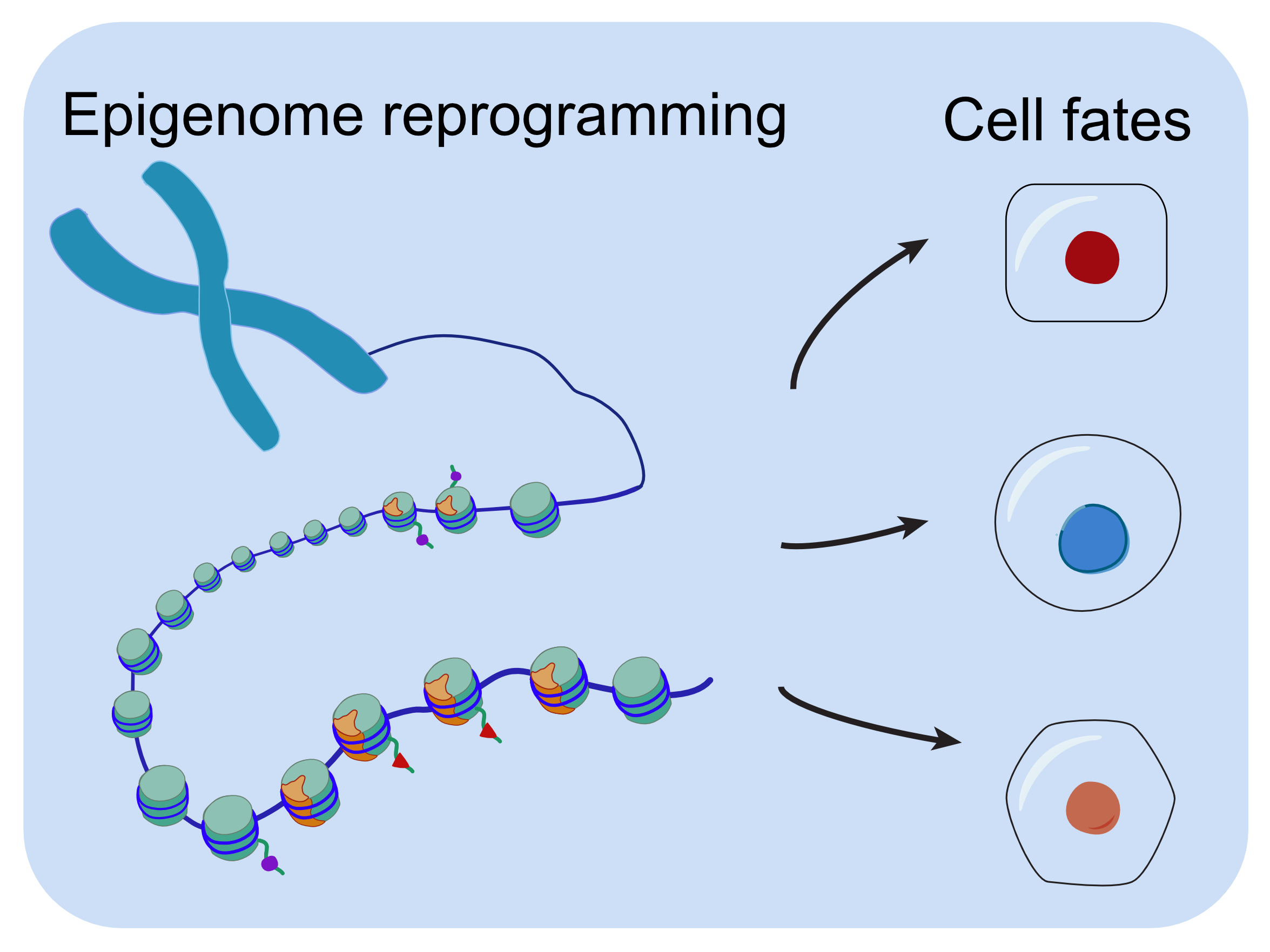 Cell fate transitions are accompanied by rewiring of gene expression networks. One critical layer of gene expression control is by epigenetic mechanisms. However, the degree to which epigenome reprogramming occurs in particular cell types and the functional role of such events are not well established. We have used recently developed genomic profiling methods such as CUT&RUN and CUT&Tag to establish spatial and temporal genomic profiles of epigenetic regulators. Currently we are studying how epigenome reprogramming facilitates development of particular lineages. Importantly, mutations of epigenetic regulators often lead to developmental disorders in humans, such as craniofacial defects. We are also interested in studying epigenetic mutation induced developmental disorders using zebrafish genetic models.
Cell fate transitions are accompanied by rewiring of gene expression networks. One critical layer of gene expression control is by epigenetic mechanisms. However, the degree to which epigenome reprogramming occurs in particular cell types and the functional role of such events are not well established. We have used recently developed genomic profiling methods such as CUT&RUN and CUT&Tag to establish spatial and temporal genomic profiles of epigenetic regulators. Currently we are studying how epigenome reprogramming facilitates development of particular lineages. Importantly, mutations of epigenetic regulators often lead to developmental disorders in humans, such as craniofacial defects. We are also interested in studying epigenetic mutation induced developmental disorders using zebrafish genetic models.
Role of repetitive elements during embryogenesis
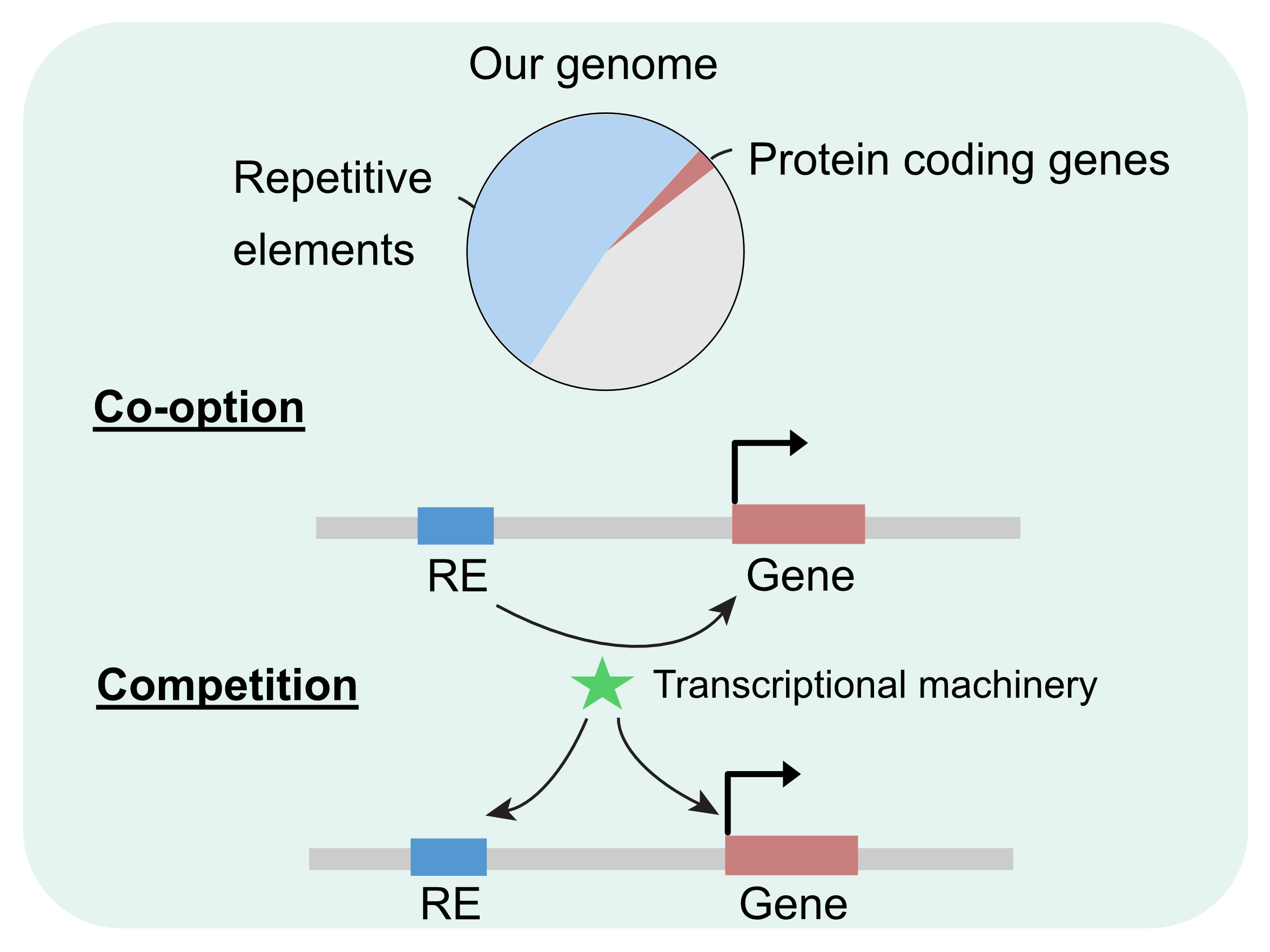
Repetitive elements account for at least half the genome in many organisms, and for decades were thought of as genomic junk with neutral or deleterious effects. Recent studies have started to appreciate the important role of repetitive elements as a key player in gene expression networks during development. REs can cooperate with transcription factors by serving as regulatory elements, on the other side, they can also compete for limited transcriptional machinery. We are particularly interested in the functional role of repetitive elements during zebrafish embryogenesis.